| Trinh Lab |
| Home | Research | Team | Publication | Download | Position | News |
 |
| Trinh lab seeks to fundamentally understand and engineer complex cellular systems with three synergistic research thrusts. Thrust 1 is to understand the principles of modular design in biological systems and develop the transformative MODCELL (Modular Cell Engineering) technology to engineer modular chassis cells for rapid development of novel microbial biocatalysts in a plug-and-play fashion for industrial biocatalysis. Thrust 2 is to understand the mechanisms to neutralize pathogens and develop the transformative ViPaRe (Virulent Pathogen Resistance) technology to effectively combat the antibiotic resistance. Thrust 3 is to understand the mechanisms of cellular robustness against environmental perturbation and develop effective defensive tools to boost cellular robustness for applications from disease prevention to novel biocatalysis. To pursue the goal, Trinh lab is interested in applying and developing both theoretical and experimental tools in interdisciplinary fields of systems and synthetic biology together with metabolic and biochemical engineering, and microbial physiology. Keywords: Metabolic Engineering, Synthetic Biology, Systems Biology, Transcriptomics, Proteomics, Metabolomics, Fluxomics, Lipiomics, Biochemical Engineering, Computational Biology, Metabolic Network Modeling, Optimization, Functional Genomics, Cell Physiology, Cell Metabolism, Viral Physiology, Cellular Robustness, CRISPR-Cas, CRISPR-Cas Antimicrobials, CRISPR-Cas Antifungals, Biotechnology, Biomanufacturing, Biofuels, Biochemicals, Biomatericals, Modular Cell Engineering, ModCell, CASPER, Escherichia coli, Bacillus coagulans, Bacillus sp., Yarrowia lipolytica, Candida albicans, Candida auris, Candida sp. |
| Topic 1. Elucidate mechanisms and develop tools to rewire cellular metabolism |
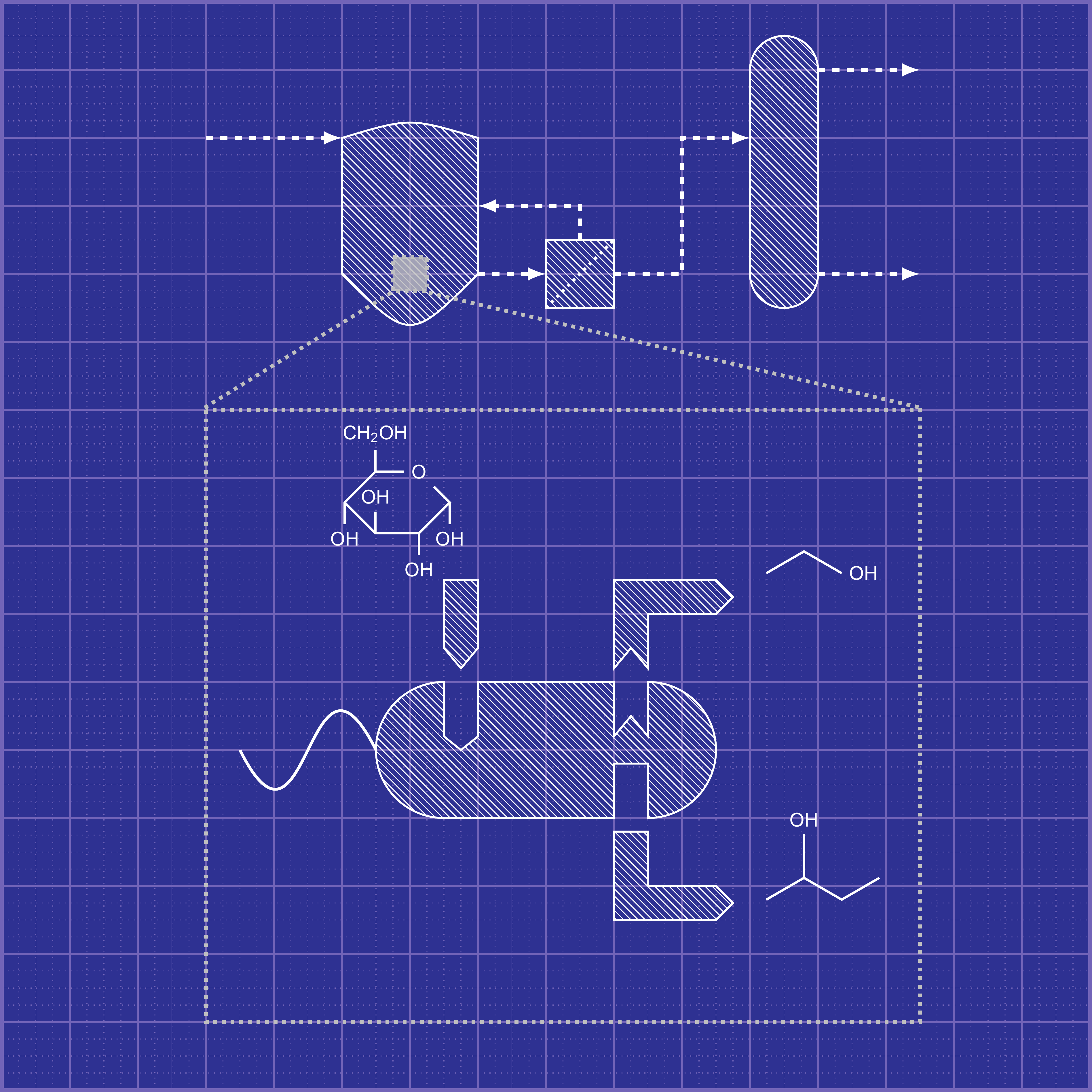
A metabolic network describing a cellular metabolism typically contains hundreds to thousands of reactions catalyzed by functional enzymes to convert substrates into precursor metabolites used for cell synthesis and other metabolites secreted to extracellular environments. These functional enzymes are directly encoded by functional and regulatory genes that determine cell phenotypes. Through metabolic network modeling, we can decompose a complex metabolic network into unique pathways, each of which contains a minimal set of enzymatic reactions supporting cell functions. Each of these independent pathways can represent physiological states of cell operation. Complete knowledge of these pathways allows the selection of cell phenotypes of interest, which establishes a basis for designing cells with optimized metabolic functionalities. The engineered cells can be rationally designed, constructed, and validated to function only according to the most efficient pathways to produce target products (e.g., chemicals, fuels, and materials). Trinh lab is particularly focused on understanding and harnessing modularity of complex biological systems for industrial biocatalysis (Figure 1). We are developing the MODCELL technology that enables rapid development of optimal production strains in a plug-and-play fashion to produce a large space of biomolecules, e.g., advanced biofuels, biochemicals, and biomaterials from renewable feedstocks or wastes (e.g., mixed plastics wastes, CO2) as well as high-valued products such as fine chemicals, secondary metabolites, and enzymes. These production strains are assembled from a universal modular (chassis) cell and different production modules based on modular cell design principles. |
|
Figure 1: Blueprint of Modular Cell Design |
| Topic 2. Understand mechanisms and develop tools to engineer cellular robustness |
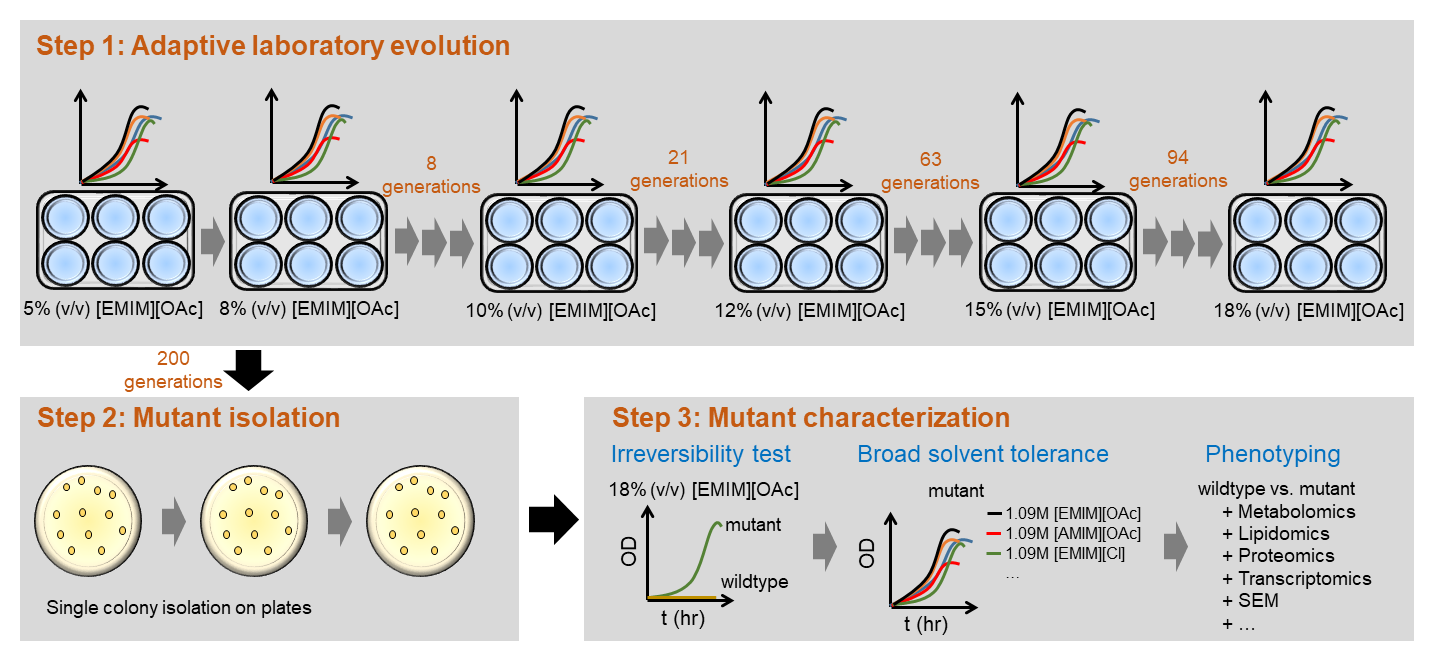
One of the challenging tasks to convert lignocellulosic biomass into biofuels is to develop robust solventogenic microorganisms that can tolerate chemical inhibitors present in the fermentation broth. These inhibitors can be unwantedly generated during pretreatment, enzymatic hydrolysis, and fermentation. Depending on the biofuel process, inhibitors can be weak organic acids (e.g., succinic acid, lactic acid, acetic acid, formic acid), furan derivatives (e.g., furfural, hydroxyl methyl furfural), phenolic compounds derived from lignin, organic solvents derived from pretreatments (e.g., ionic liquids), and/or fermentative products synthesized by microorganisms (e.g., ethanol, (iso)butanol, esters). These inhibitors cause detrimental effect on biocatalyst performance by decreasing both cell growth and solvent production. It is difficult to metabolically engineer microorganisms resistant to these chemical inhibitors because the genotype and phenotype links resulting in chemical resistance in microorganisms are complex and very often unknown. |
Figure 2: Pipeline of adaptive laboratory evolution and genome-wide analysis to understand and optimze cellular robustness |
To engineer cellular robustness, Trinh lab has applied and developed various genome engineering tools. One developed tool is the gene swapping and amplification that is designed to transfer novel phenotypes from not-well-characterized microorganisms exhibiting high resistance to chemical inhibitors to an engineered host. The other approach is based on the classical but effective adaptive laboratory evolution (Figure 2). In parallel to the engineering efforts, Trinh lab is also interested in developing and applying novel omics tools to elucidate the genetics and underlying mechanisms enable cellular robustness through systems-wide analysis. |
Funding sources: The U.S. National Science Foundation; The U.S. Department of Energy. |
| Topic 3. Elucidate design principles and develop tools to engineer synthetic pathways |
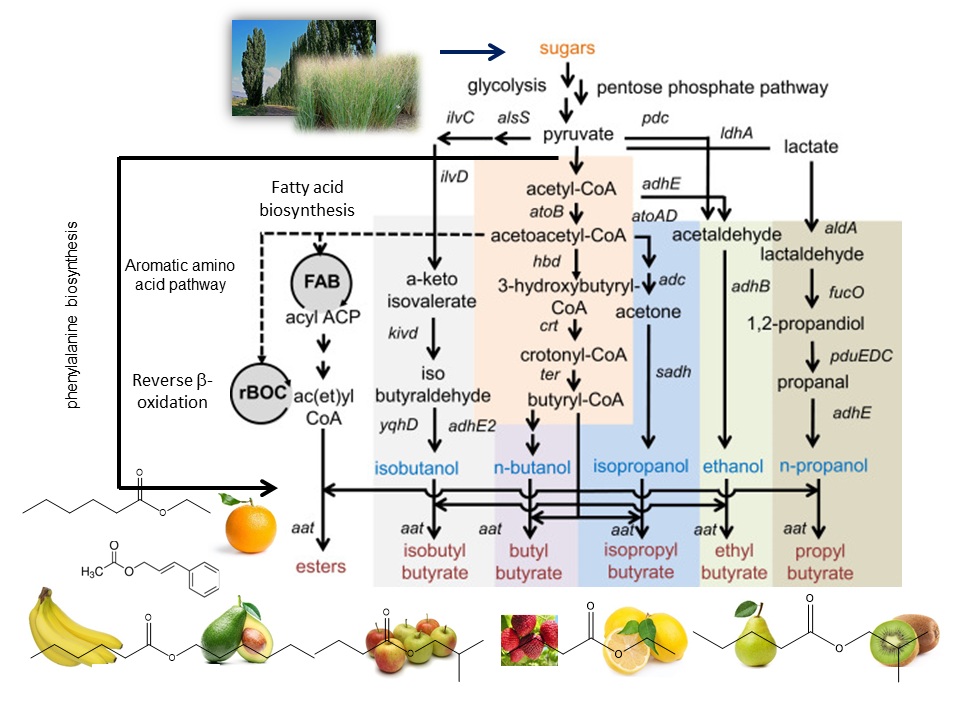
Developing an efficient and robust whole-cell biocatalyst to produce a target chemical requires the recruitment of heterologous genes to constitute a synthetic metabolic pathway in the microorganism. Even though advances in the recombinant DNA technology have developed powerful and convenient molecular biology tools to transfer genes among species, it is still a challenging task to optimize the operation of synthetic metabolic pathways in a microorganism due to imbalanced metabolic fluxes not only in the synthetic metabolic pathway but also in other associated pathways. This imbalance results in the accumulation of intermediates that are toxic to cells, which inhibits cell growth and decreases production of a target metabolite. Trinh lab is interested in developing novel prototypes to efficiently and rapidly design and optimize the performance of synthetic operons that can dynamically control fluxes of synthetic and native metabolic pathways for enhanced product formation in engineered cells. We are currently exploring a large space of novel pathways for combinatorial microbial biosynthesis of esters that can be used as flavors, fragrances, solvents, and biofuels (Figure 3). |
Figure 3: Metabolic pathway platform for biosynthesis of butyrate esters |
Funding sources: The U.S. National Science Foundation; The U.S. Department of Energy. |
| Topic 4. Understand mechanisms and develop tools for directed metabolic pathway evolution |
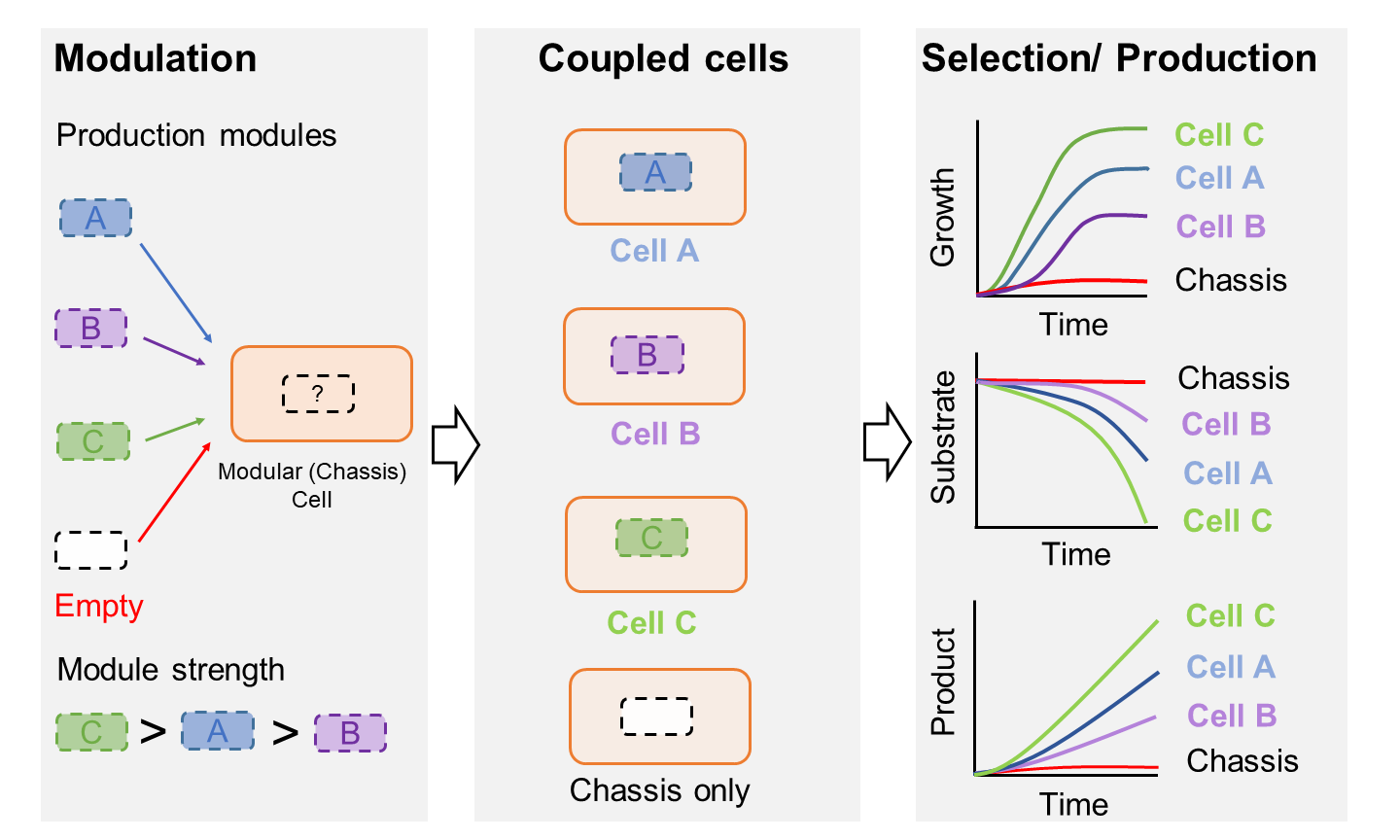
An optimized cell designed to couple cell growth and operation of a target pathway can be utilized as an useful host to conduct the in vivo metabolic pathway evolution because cell growth is inhibited without the functioning of the target pathway. The selection basis for the molecular pathway evolution is growth phenotype which is easy to implement. |
Figure 4: Directed metabolic pathway evolution using ModCell design |
We are exploring this approach using our MODCELL technology to improve titers, yields, and productivities for microbial production of biofuels, biochemicals, secondary metabolites, and enzymes. With this approach, the metabolic pathway evolution can be implemented by replacing the enzyme of a reaction within the target pathway with a potential candidate from a library of mutated enzymes or from different microorganisms that have similar or related functions (Figure 4). Only cells containing complementary enzymes can support cell growth. The candidate can be selected by using advanced cell-culturing techniques such as a cytostat, turbidostat, and/or CO2-stat. With these selection methods, only host cells containing enzymes with high activities are enriched. In parallel, we are highly interested in better understanding the underlying mechanisms of the molecular pathway evolution resulting in desirable phenotypes by using systems-biology tools and hence discovering novel genotype-phenotype links to guide the inverse metabolic engineering. Funding sources: The U.S. National Science Foundation; The U.S. Department of Energy. |
| Topic 5. Understand mechanisms and develop tools to tackle antibiotic resistance |
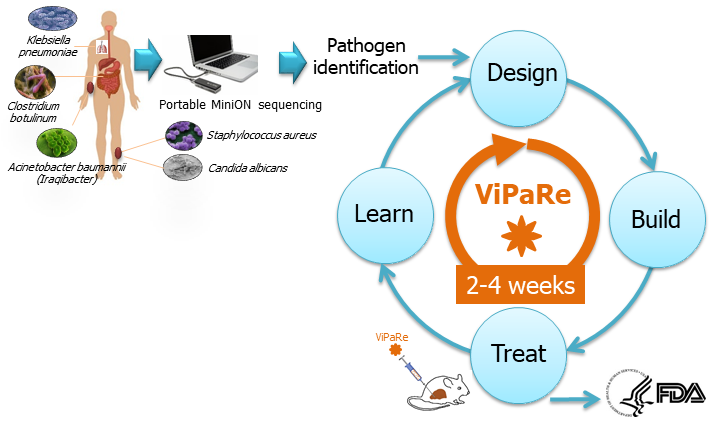
In the war against virulent pathogens, defensive measures must be developed quickly and be readily modifiable to counter rapid and unpredictable evolution of resistance. The current paradigm relies on small molecule discovery technologies that are unable to scale to production fast enough to counter pathogens that can rapidly adapt to neutralize existing treatments. To address the challenge, we seek to develop the ViPaRe (Virulent Pathogen Resistance) technology capable of neutralizing rapidly evolving and resistant infectious pathogens (e.g, Staphylococcus aureus, Candida albicans, Candida auris, Escherichia coli ) in their microbiomes using the CRISPR-based gene editing technology (Figure 5). |
Figure 5: ViPaRe technology |
Funding sources: The U.S. Department of Defense. |
| Updated on 01.24.2025 |  |